| Citation: | Haichao Fang, Qiwen Sun. THE DYNAMICS OF GENE TRANSCRIPTION INDUCED BY VARIATION IN TRANSCRIPTION KINETICS[J]. Journal of Applied Analysis & Computation, 2023, 13(5): 2955-2971. doi: 10.11948/20230072 |
THE DYNAMICS OF GENE TRANSCRIPTION INDUCED BY VARIATION IN TRANSCRIPTION KINETICS
-
Abstract
In single cells, the process of gene transcription generally demonstrates complicated and stochastic behaviors. The stochasticity of transcription brings about large variations in the number of mRNA molecules, even in a homogeneous intracellular environment. Randomly switching between periods of active and inactive gene expression is considered to be the main cause of the high variation of the mRNA distributions. Many studies have revealed that the transcription system will enter a steady state after several transcription cycles in the last three decades. Changes in the intracellular or intercellular environment give rise to changes in transcription parameters, resulting in perturbations of a homeostatic state. In this paper, we mainly studied the dynamic behaviors of the mean mRNA level and the noise following the occurrence of the variation in transcription kinetics. We defined three quantities that are used to determine the monotonicity of the average transcription level. When the mean level is not monotonous, the value may reach the potential thresholds, thereby changing the fate of cells. This is extremely significant for researching gene expression regulation.
-
-
References
[1] A. Bar-Even, J. Paulsson, N. Maheshri, et al., Noise in protein expression scales with natural protein abundance, Nature Genetics, 2006, 38, 636–643. doi: 10.1038/ng1807 [2] Z. Cao and R. Grima, Analytical distributions for detailed models of stochastic gene expression in eukaryotic cells, Proceedings of the National Academy of Sciences of the United States of America, 2020, 117, 4682–4692. [3] Z. Cao and R. Grima, Linear mapping approximation of gene regulatory networks with stochastic dynamics, Nature Communications, 2018, 9, 3305. doi: 10.1038/s41467-018-05822-0 [4] Z. Cao, F. Qian and R. Grima, Neural network aided approximation and parameter inference of non-Markovian models of gene expression, Nature Communications, 2021, 12, 2618. doi: 10.1038/s41467-021-22919-1 [5] P. Caveney, S. Norred, C. Chin, et al., Resource sharing controls gene expression bursting, ACS Synthetic Biology, 2017, 6(2), 334–343. doi: 10.1021/acssynbio.6b00189 [6] V. Chauhan, M. Bahrudeen, C. Palma, et al., Analytical kinetic model of native tandem promoters in E. coli, PLoS Computational Biology, 2022, 18, e1009824. doi: 10.1371/journal.pcbi.1009824 [7] L. Chen, Y. Lin, D. Gallegos, et al., Enhancer histone acetylation modulates transcriptional bursting dynamics of neuronal activity-inducible genes, Cell Reports, 2019, 26(6), 1174–1188. [8] A. Corrigan, E. Tunnacliffe, D. Cannon and J. Chubb, A continuum model of transcriptional bursting, eLife, 2016, 5, e13051. doi: 10.7554/eLife.13051 [9] R. Dar, B. Razooky, L. Weinberger, C. Cox and M. Simpson, The low noise limit in gene expression, PLoS ONE, 2015, 10, e0140969. doi: 10.1371/journal.pone.0140969 [10] H. Fraser, Cell-cycle regulated transcription assocates with DNA replication timing in yeast and human, Genome Biology, 2013, 14, R111. doi: 10.1186/gb-2013-14-10-r111 [11] A. Gjuvsland, E. Plahte and S. Omholt, Threshold-dominated regulation hides genetic variation in gene expression networks, BMC Systems Biology, 2007, 1, 57. doi: 10.1186/1752-0509-1-57 [12] M. Guo, Y. Du, J. Gokey, et al., Single cell RNA analysis identifies cellular heterogeneity and adaptive responses of the lung at birth, Nature Communications, 2019, 10, 37. doi: 10.1038/s41467-018-07770-1 [13] A. Hansen and E. O'Shea, cis determinants of promoter threshold and activation timescale, Cell Reports, 2015, 12, 1226–1233. doi: 10.1016/j.celrep.2015.07.035 [14] C. Jia, Simplification of Markov chains with infinite state space and the mathematical theory of random gene expression bursts, Physical Review E, 2017, 96, 032402. doi: 10.1103/PhysRevE.96.032402 [15] C. Jia, Kinetic foundation of the zero-inflated negative binomial model for single-cell RNA sequencing data, SIAM Journal on Applied Mathematics, 2020, 80, 1336–1355. doi: 10.1137/19M1253198 [16] C. Jia and R. Grima, Frequency domain analysis of fluctuations of mRNA and protein copy numbers within a cell lineage: Theory and experimental validation, Physical Review X, 2021, 11, 021032. [17] F. Jiao, G. Lin and J. Yu, Approximating gene transcription dynamics using steady-state formulas, Physical Review E, 2021, 104, 014401. [18] F. Jiao and M. Tang, Quantification of transcription noise's impact on cell fate commitment with digital resolutions, Bioinformatics, 2022, 38, 3062–3069. doi: 10.1093/bioinformatics/btac277 [19] F. Jiao, M. Tang and J. Yu, Distribution profiles and their dynamic transition in stochastic gene transcription, Journal of Differential Equations, 2013, 254, 3307–3328. doi: 10.1016/j.jde.2013.01.019 [20] F. Jiao and C. Zhu, Regulation of gene activation by competitive cross talking pathways, Biophysical Journal, 2020, 119, 1204–1214. doi: 10.1016/j.bpj.2020.08.011 [21] B. Kaufmann and A. van Oudenaarden, Stochastic gene expression: from single molecules to the proteome, Current Opinion in Genetics & Development, 2007, 17, 107–112. [22] M. Ko, A stochastic model for gene induction, Journal of Theoretical Biology, 1991, 153, 181–194. doi: 10.1016/S0022-5193(05)80421-7 [23] J. Kuang, M. Tang and J. Yu, The mean and noise of protein numbers in stochastic gene expression, Journal of Mathematical Biology, 2013, 67, 261–291. doi: 10.1007/s00285-012-0551-8 [24] A. Larsson, P. Johnsson, M. Hagemann-Jensen, et al., Genomic encoding of transcriptional burst kinetics, Nature, 2019, 565, 251–254. doi: 10.1038/s41586-018-0836-1 [25] J. Little, Threshold effects in gene regulation: When some is not enough, Proceedings of the National Academy of Sciences of the United States of America, 2005, 102, 5310–5311. [26] N. Maheshri and E. O'Shea, Living with noisy genes: how cells function reliably with inherent variability in gene expression, Annual Review of Biophysics and Biomolecular Structure, 2007, 36, 413–434. doi: 10.1146/annurev.biophys.36.040306.132705 [27] S. Marguerat and J. Bälher, Coordinating genome expression with cell size, Trends in Genetics, 2012, 28, 560–565. doi: 10.1016/j.tig.2012.07.003 [28] N. Molina, D. M. Suter, R. Cannavo, et al., Stimulus-induced modulation of transcriptional bursting in a single mammalian gene, Proceedings of the National Academy of Sciences of the United States of America, 2013, 110, 20563–20568. [29] S. Mukherji, M. Ebert, G. Zheng, et al., MicroRNAs can generate thresholds in target gene expression, Nat Genet, 2011, 43, 854–859. doi: 10.1038/ng.905 [30] B. Munsky, G. Neuert and A. van Oudenaarden, Using gene expression noise to understand gene regulation, Science, 2012, 336, 183–187. doi: 10.1126/science.1216379 [31] G. Neuert, B. Munsky, R. Tan, et al., Systematic identification of signal-activated stochastic gene regulation, Science, 2013, 339, 584–587. doi: 10.1126/science.1231456 [32] O. Padovan-Merhar, G. Nair, A. Biaesch, et al., Single mammalian cells compensate for differences in cellular volume and DNA copy number through independent global transcriptional mechanisms, Molecular Cell, 2015, 58, 339–352. doi: 10.1016/j.molcel.2015.03.005 [33] M. Prajapat and A. Ribeiro, Added value of autoregulation and multi-step kinetics of transcription initiation, Royal Society Open Science, 2018, 5, 181170. doi: 10.1098/rsos.181170 [34] A. Raj, C. Peskin, D. Tranchina, et al., Stochastic mRNA synthesis in mammalian cells, PLoS Biology, 2006, 4, 1707–1719. [35] S. Skinner, H. Xu, S. Nagarkar-Jaiswal, et al., Single-cell analysis of transcription kinetics across the cell cycle, eLife, 2016, 5, e12175. doi: 10.7554/eLife.12175 [36] Q. Sun, Z. Cai and C. Zhu, A novel dynamical regulation of mRNA distribution by cross-talking pathways, Mathematics, 2022, 10, 1515. doi: 10.3390/math10091515 [37] Q. Sun, F. Jiao, G. Lin, et al., The nonlinear dynamics and fluctuations of mRNA levels in cell cycle coupled transcription, PLoS Computational Biology, 2019, 15, e1007017. doi: 10.1371/journal.pcbi.1007017 [38] Q. Sun, F. Jiao and J. Yu, The dynamics of gene transcription with a periodic synthesis rate, Nonlinear Dynamics, 2021, 104, 4477–4492. doi: 10.1007/s11071-021-06569-y [39] M. Tang, The mean and noise of stochastic gene transcription, Journal of Theoretical Biology, 2008, 253, 271–280. doi: 10.1016/j.jtbi.2008.03.023 [40] T. Trcek, D. Larson, A. Moldón, et al., Single-molecule mRNA decay measurements reveal promoter-regulated mRNA stability in yeast, Cell, 2011, 147, 1484–1497. doi: 10.1016/j.cell.2011.11.051 [41] Y. Voichek, R. Bar-Ziv and N. Barkai, Expression homeostasis during DNA replication, Science, 2016, 351, 1087–1090. doi: 10.1126/science.aad1162 [42] J. Wang, S. Zhang, H. Lu and H. Xu, Differential regulation of alternative promoters emerges from unified kinetics of enhancer-promoter interaction, Nature Communications, 2022, 13, 2714. doi: 10.1038/s41467-022-30315-6 [43] H. Xu, S. Skinner, A. Sokac and I. Golding, Stochastic kinetics of nascent RNA, Physical Review Letter, 2016, 117, 128101. doi: 10.1103/PhysRevLett.117.128101 [44] J. Yu, Q. Sun and M. Tang, The nonlinear dynamics and fluctuations of mRNA levels in cross-talking pathway activated transcription, Journal of Theoretical Biology, 2014, 363, 223–234. doi: 10.1016/j.jtbi.2014.08.024 [45] J. Zhang and T. Zhou, Markovian approaches to modeling intracellular reaction processes with molecular memory, Proceedings of the National Academy of Sciences of the United States of America, 2019, 116, 23542–23550. [46] C. Zhu, Z. Chen and Q. Sun, Stochastic transcription with alterable synthesis rates, Mathematics, 2022, 10, 2189. doi: 10.3390/math10132189 [47] C. Zopf, K. Quinn, J. Zeidman and N. Maheshri, Cell-cycle dependence of transcription dominates noise in gene expression, PLoS Computational Biology, 2013, 9, e1003161. doi: 10.1371/journal.pcbi.1003161 -
-
-
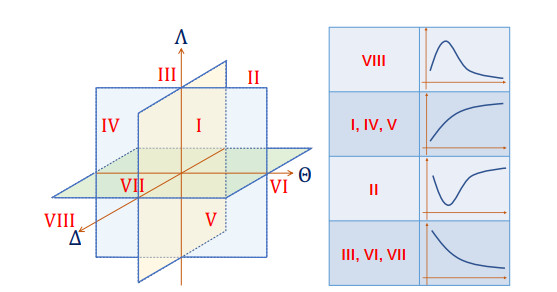
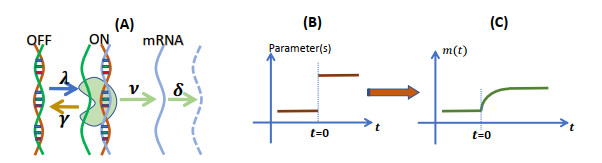
Figure 1.
Stochastic gene transcription induced by parameter changes. (A) The promoter is activated by binding transcription factors to the regulation region, and transcription starts when RNA polymerase binds to a promoter, and then moves along the template and produces an RNA chain. (B) Transcription is regulated frequently by RNA polymerase and other regulatory factors, leading to variations in transcription kinetics. (C) The variation in parameters enables the transcription system to deviate from the original equilibrium state.
-
Figure 2.
The planes
$ \Delta =0 $ $ \Theta=0 $ $ \Lambda=0 $ $ m(t) $ $ (\Delta, \Theta, \Lambda) $ -
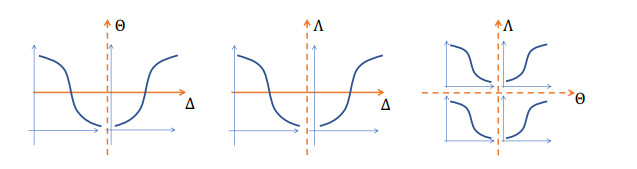
Figure 3.
The dynamic behaviors of
$ m(t) $ $ (\Delta, \Theta, \Lambda) $ $ m(t) $ $ (\Delta, \Theta, \Lambda) $ $ (\Delta, \Theta, \Lambda) $ $ \Lambda $ $ (0, 0, 0) $ -
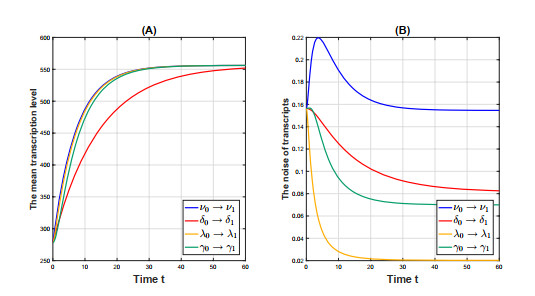
Figure 4.
The temporal profiles of the mean transcription level
$ m(t) $ $ \eta^2(t) $ $ m(t) $ $ \eta^2(t) $ -
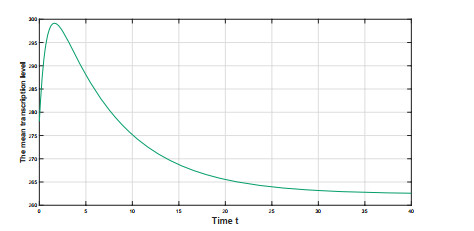
Figure 5.
The temporal profile of the mean transcription level
$ m(t) $ $ (\Delta, \Theta, \Lambda)=(814.3, -0.1804, -58.9184)\in{\rm VIII} $ $ m(t) $ $ 278.1 $ $ 299.1 $ $ t=1.6 $ $ 262.4 $



 DownLoad:
DownLoad: